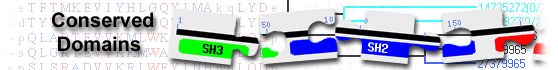integral membrane C-type cytochrome biogenesis protein DipZ [Mycobacterium tuberculosis H37Rv]
cytochrome c biogenesis protein DipZ( domain architecture ID 14298428)
integral membrane cytochrome c biogenesis protein DipZ, or DsbD, is involved in disulfide bond formation of target proteins by transporting electrons from cytoplasmic thioredoxin to DsbC and DsbG
List of domain hits

| Name | Accession | Description | Interval | E-value | ||||
| TlpA_like_DipZ_like | cd03012 | TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a ... |
401-528 | 5.45e-74 | ||||
TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a TlpA-like TRX domain. Some members show domain architectures similar to that of E. coli DipZ protein (also known as DsbD). The only eukaryotic members of the TlpA family belong to this subfamily. TlpA is a disulfide reductase known to have a crucial role in the biogenesis of cytochrome aa3. : Pssm-ID: 239310 [Multi-domain] Cd Length: 126 Bit Score: 234.51 E-value: 5.45e-74
|
||||||||
| Thioredoxin_10 | pfam17991 | Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in ... |
568-695 | 5.99e-41 | ||||
Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in Rv2874 in the pathogenic bacterium Mycobacterium tuberculosis. Structure analysis of Rv2874-C shows the presence of a C-terminal domain formed by the 128 residues Thr568-Gly695. These residues form a jelly-roll structure in which two antiparallel beta-sheets sandwich a hydrophobic core. This domain is combined with a second domain with a carbohydrate-binding module (CBM) fold. : Pssm-ID: 465607 Cd Length: 142 Bit Score: 146.19 E-value: 5.99e-41
|
||||||||
| CcdA | COG0785 | Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational ... |
115-321 | 5.56e-38 | ||||
Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational modification, protein turnover, chaperones]; : Pssm-ID: 440548 [Multi-domain] Cd Length: 193 Bit Score: 139.59 E-value: 5.56e-38
|
||||||||
| Name | Accession | Description | Interval | E-value | ||||
| TlpA_like_DipZ_like | cd03012 | TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a ... |
401-528 | 5.45e-74 | ||||
TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a TlpA-like TRX domain. Some members show domain architectures similar to that of E. coli DipZ protein (also known as DsbD). The only eukaryotic members of the TlpA family belong to this subfamily. TlpA is a disulfide reductase known to have a crucial role in the biogenesis of cytochrome aa3. Pssm-ID: 239310 [Multi-domain] Cd Length: 126 Bit Score: 234.51 E-value: 5.45e-74
|
||||||||
| Thioredoxin_10 | pfam17991 | Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in ... |
568-695 | 5.99e-41 | ||||
Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in Rv2874 in the pathogenic bacterium Mycobacterium tuberculosis. Structure analysis of Rv2874-C shows the presence of a C-terminal domain formed by the 128 residues Thr568-Gly695. These residues form a jelly-roll structure in which two antiparallel beta-sheets sandwich a hydrophobic core. This domain is combined with a second domain with a carbohydrate-binding module (CBM) fold. Pssm-ID: 465607 Cd Length: 142 Bit Score: 146.19 E-value: 5.99e-41
|
||||||||
| CcdA | COG0785 | Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational ... |
115-321 | 5.56e-38 | ||||
Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 440548 [Multi-domain] Cd Length: 193 Bit Score: 139.59 E-value: 5.56e-38
|
||||||||
| TrxA | COG0526 | Thiol-disulfide isomerase or thioredoxin [Posttranslational modification, protein turnover, ... |
399-542 | 3.28e-31 | ||||
Thiol-disulfide isomerase or thioredoxin [Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 440292 [Multi-domain] Cd Length: 139 Bit Score: 118.64 E-value: 3.28e-31
|
||||||||
| DsbD | pfam02683 | Cytochrome C biogenesis protein transmembrane region; This family consists of the ... |
122-340 | 4.36e-18 | ||||
Cytochrome C biogenesis protein transmembrane region; This family consists of the transmembrane (i.e. non-catalytic) region of Cytochrome C biogenesis proteins also known as disulphide interchange proteins. These proteins posses a protein disulphide isomerase like domain that is not found within the aligned region of this family. Pssm-ID: 280792 [Multi-domain] Cd Length: 213 Bit Score: 83.61 E-value: 4.36e-18
|
||||||||
| PLN02919 | PLN02919 | haloacid dehalogenase-like hydrolase family protein |
392-528 | 3.49e-16 | ||||
haloacid dehalogenase-like hydrolase family protein Pssm-ID: 215497 [Multi-domain] Cd Length: 1057 Bit Score: 82.98 E-value: 3.49e-16
|
||||||||
| AhpC-TSA | pfam00578 | AhpC/TSA family; This family contains proteins related to alkyl hydroperoxide reductase (AhpC) ... |
400-523 | 6.47e-12 | ||||
AhpC/TSA family; This family contains proteins related to alkyl hydroperoxide reductase (AhpC) and thiol specific antioxidant (TSA). Pssm-ID: 425763 [Multi-domain] Cd Length: 124 Bit Score: 63.01 E-value: 6.47e-12
|
||||||||
| Name | Accession | Description | Interval | E-value | ||||||
| TlpA_like_DipZ_like | cd03012 | TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a ... |
401-528 | 5.45e-74 | ||||||
TlpA-like family, DipZ-like subfamily; composed uncharacterized proteins containing a TlpA-like TRX domain. Some members show domain architectures similar to that of E. coli DipZ protein (also known as DsbD). The only eukaryotic members of the TlpA family belong to this subfamily. TlpA is a disulfide reductase known to have a crucial role in the biogenesis of cytochrome aa3. Pssm-ID: 239310 [Multi-domain] Cd Length: 126 Bit Score: 234.51 E-value: 5.45e-74
|
||||||||||
| Thioredoxin_10 | pfam17991 | Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in ... |
568-695 | 5.99e-41 | ||||||
Thioredoxin like C-terminal domain; This is the C-terminal thioredoxin like domain found in Rv2874 in the pathogenic bacterium Mycobacterium tuberculosis. Structure analysis of Rv2874-C shows the presence of a C-terminal domain formed by the 128 residues Thr568-Gly695. These residues form a jelly-roll structure in which two antiparallel beta-sheets sandwich a hydrophobic core. This domain is combined with a second domain with a carbohydrate-binding module (CBM) fold. Pssm-ID: 465607 Cd Length: 142 Bit Score: 146.19 E-value: 5.99e-41
|
||||||||||
| CcdA | COG0785 | Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational ... |
115-321 | 5.56e-38 | ||||||
Cytochrome c biogenesis protein CcdA [Energy production and conversion, Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 440548 [Multi-domain] Cd Length: 193 Bit Score: 139.59 E-value: 5.56e-38
|
||||||||||
| TlpA_like_family | cd02966 | TlpA-like family; composed of TlpA, ResA, DsbE and similar proteins. TlpA, ResA and DsbE are ... |
410-526 | 3.57e-34 | ||||||
TlpA-like family; composed of TlpA, ResA, DsbE and similar proteins. TlpA, ResA and DsbE are bacterial protein disulfide reductases with important roles in cytochrome maturation. They are membrane-anchored proteins with a soluble TRX domain containing a CXXC motif located in the periplasm. The TRX domains of this family contain an insert, approximately 25 residues in length, which correspond to an extra alpha helix and a beta strand when compared with TRX. TlpA catalyzes an essential reaction in the biogenesis of cytochrome aa3, while ResA and DsbE are essential proteins in cytochrome c maturation. Also included in this family are proteins containing a TlpA-like TRX domain with domain architectures similar to E. coli DipZ protein, and the N-terminal TRX domain of PilB protein from Neisseria which acts as a disulfide reductase that can recylce methionine sulfoxide reductases. Pssm-ID: 239264 [Multi-domain] Cd Length: 116 Bit Score: 126.20 E-value: 3.57e-34
|
||||||||||
| TrxA | COG0526 | Thiol-disulfide isomerase or thioredoxin [Posttranslational modification, protein turnover, ... |
399-542 | 3.28e-31 | ||||||
Thiol-disulfide isomerase or thioredoxin [Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 440292 [Multi-domain] Cd Length: 139 Bit Score: 118.64 E-value: 3.28e-31
|
||||||||||
| Bcp | COG1225 | Peroxiredoxin [Posttranslational modification, protein turnover, chaperones]; |
400-545 | 6.19e-25 | ||||||
Peroxiredoxin [Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 440838 [Multi-domain] Cd Length: 136 Bit Score: 100.71 E-value: 6.19e-25
|
||||||||||
| DsbD | pfam02683 | Cytochrome C biogenesis protein transmembrane region; This family consists of the ... |
122-340 | 4.36e-18 | ||||||
Cytochrome C biogenesis protein transmembrane region; This family consists of the transmembrane (i.e. non-catalytic) region of Cytochrome C biogenesis proteins also known as disulphide interchange proteins. These proteins posses a protein disulphide isomerase like domain that is not found within the aligned region of this family. Pssm-ID: 280792 [Multi-domain] Cd Length: 213 Bit Score: 83.61 E-value: 4.36e-18
|
||||||||||
| PLN02919 | PLN02919 | haloacid dehalogenase-like hydrolase family protein |
392-528 | 3.49e-16 | ||||||
haloacid dehalogenase-like hydrolase family protein Pssm-ID: 215497 [Multi-domain] Cd Length: 1057 Bit Score: 82.98 E-value: 3.49e-16
|
||||||||||
| PRK03147 | PRK03147 | thiol-disulfide oxidoreductase ResA; |
414-546 | 1.58e-13 | ||||||
thiol-disulfide oxidoreductase ResA; Pssm-ID: 179545 [Multi-domain] Cd Length: 173 Bit Score: 69.26 E-value: 1.58e-13
|
||||||||||
| DsbD | COG4232 | Thiol:disulfide interchange protein DsbD [Posttranslational modification, protein turnover, ... |
119-441 | 5.84e-12 | ||||||
Thiol:disulfide interchange protein DsbD [Posttranslational modification, protein turnover, chaperones]; Pssm-ID: 443376 [Multi-domain] Cd Length: 416 Bit Score: 68.29 E-value: 5.84e-12
|
||||||||||
| AhpC-TSA | pfam00578 | AhpC/TSA family; This family contains proteins related to alkyl hydroperoxide reductase (AhpC) ... |
400-523 | 6.47e-12 | ||||||
AhpC/TSA family; This family contains proteins related to alkyl hydroperoxide reductase (AhpC) and thiol specific antioxidant (TSA). Pssm-ID: 425763 [Multi-domain] Cd Length: 124 Bit Score: 63.01 E-value: 6.47e-12
|
||||||||||
| TauE | COG0730 | Sulfite exporter TauE/SafE/YfcA and related permeases, UPF0721 family [Inorganic ion transport ... |
111-342 | 2.79e-07 | ||||||
Sulfite exporter TauE/SafE/YfcA and related permeases, UPF0721 family [Inorganic ion transport and metabolism]; Pssm-ID: 440494 Cd Length: 250 Bit Score: 52.12 E-value: 2.79e-07
|
||||||||||
| Redoxin | pfam08534 | Redoxin; This family of redoxins includes peroxiredoxin, thioredoxin and glutaredoxin proteins. |
399-527 | 1.59e-06 | ||||||
Redoxin; This family of redoxins includes peroxiredoxin, thioredoxin and glutaredoxin proteins. Pssm-ID: 400717 [Multi-domain] Cd Length: 148 Bit Score: 48.13 E-value: 1.59e-06
|
||||||||||
| Gdt1 | COG2119 | Putative Ca2+/H+ antiporter, TMEM165/GDT1 family [General function prediction only]; |
168-342 | 2.86e-05 | ||||||
Putative Ca2+/H+ antiporter, TMEM165/GDT1 family [General function prediction only]; Pssm-ID: 441722 [Multi-domain] Cd Length: 192 Bit Score: 45.58 E-value: 2.86e-05
|
||||||||||
| TauE | pfam01925 | Sulfite exporter TauE/SafE; This is a family of integral membrane proteins where the alignment ... |
118-336 | 6.45e-05 | ||||||
Sulfite exporter TauE/SafE; This is a family of integral membrane proteins where the alignment appears to contain two duplicated modules of three transmembrane helices. The proteins are involved in the transport of anions across the cytoplasmic membrane during taurine metabolism as an exporter of sulfoacetate. This family used to be known as DUF81. Pssm-ID: 460386 Cd Length: 235 Bit Score: 44.88 E-value: 6.45e-05
|
||||||||||
| Thioredoxin_8 | pfam13905 | Thioredoxin-like; Thioredoxins are small enzymes that participate in redox reactions, via the ... |
425-520 | 7.03e-05 | ||||||
Thioredoxin-like; Thioredoxins are small enzymes that participate in redox reactions, via the reversible oxidation of an active centre disulfide bond. Pssm-ID: 464033 [Multi-domain] Cd Length: 95 Bit Score: 42.29 E-value: 7.03e-05
|
||||||||||
| RhtB | COG1280 | Threonine/homoserine/homoserine lactone efflux protein [Amino acid transport and metabolism]; |
165-341 | 2.25e-04 | ||||||
Threonine/homoserine/homoserine lactone efflux protein [Amino acid transport and metabolism]; Pssm-ID: 440891 Cd Length: 205 Bit Score: 42.90 E-value: 2.25e-04
|
||||||||||
| PRX_like1 | cd02969 | Peroxiredoxin (PRX)-like 1 family; hypothetical proteins that show sequence similarity to PRXs. ... |
400-521 | 5.98e-04 | ||||||
Peroxiredoxin (PRX)-like 1 family; hypothetical proteins that show sequence similarity to PRXs. Members of this group contain a conserved cysteine that aligns to the first cysteine in the CXXC motif of TRX. This does not correspond to the peroxidatic cysteine found in PRXs, which aligns to the second cysteine in the CXXC motif of TRX. In addition, these proteins do not contain the other two conserved residues of the catalytic triad of PRX. PRXs confer a protective antioxidant role in cells through their peroxidase activity in which hydrogen peroxide, peroxynitrate, and organic hydroperoxides are reduced and detoxified using reducing equivalents derived from either thioredoxin, glutathione, trypanothione and AhpF. Pssm-ID: 239267 [Multi-domain] Cd Length: 171 Bit Score: 41.07 E-value: 5.98e-04
|
||||||||||
| TgpA_N | pfam11992 | TgpA N-terminal domain; This domain can be found at the N terminus of TgpA from Pseudomonas ... |
189-376 | 6.99e-03 | ||||||
TgpA N-terminal domain; This domain can be found at the N terminus of TgpA from Pseudomonas aeruginosa. TgpA is a transglutaminase that plays a critical role in the viability of Pseudomonas aeruginosa. This domain is composed of 5 transmembrane helices. Pssm-ID: 463423 Cd Length: 336 Bit Score: 39.34 E-value: 6.99e-03
|
||||||||||
| Blast search parameters | ||||
|