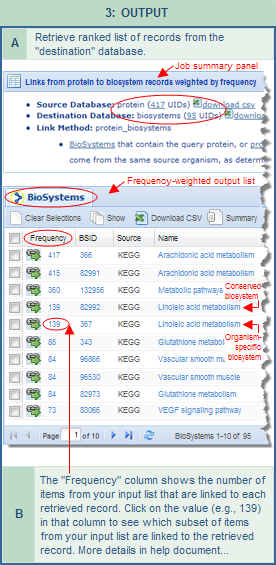
| direct input of UID list into FLink | search an Entrez database and then link to FLink | maximum input |
 


One method of using  FLink is to input a list of unique identifiers (UIDs) from any one of the supported databases directly into the tool. Valid UIDs are integers only, and the current range of acceptable is 0 to 4294967295. The input list should contain no alphanumeric combinations or other non-integer characters. For example, the GI numbers of sequence records are allowable but the accession numbers are not. As another example, if your input UIDs are for biosystem records, they should be formatted as digits only, e.g., 82991 not bsid82991. (Tip: the Batch Entrez tool can be used to screen your input list to identify obsolete and otherwise invalid IDs, if desired.) FLink is to input a list of unique identifiers (UIDs) from any one of the supported databases directly into the tool. Valid UIDs are integers only, and the current range of acceptable is 0 to 4294967295. The input list should contain no alphanumeric combinations or other non-integer characters. For example, the GI numbers of sequence records are allowable but the accession numbers are not. As another example, if your input UIDs are for biosystem records, they should be formatted as digits only, e.g., 82991 not bsid82991. (Tip: the Batch Entrez tool can be used to screen your input list to identify obsolete and otherwise invalid IDs, if desired.)
|
| |
Tip: You can use Batch Entrez, if desired, to screen a UID list and identify obsolete or otherwise invalid UIDs. Be sure to select the appropriate database from the pull-down menu at the top of the page, then "Browse" to find the file of UIDs you'd like to upload, and press "Retrieve." Batch Entrez will then display a report summarizing the data it found in your input file, such as: (1) number of lines that were present in the file (there should be one UID per line); (2) rejected lines (indicating how many invalid UIDs were detected); (3) removed duplicates (indicating how many duplicate UIDs were detected/removed); (4) passed to Entrez (indicating how many of the UIDs from your list are valid and will therefore be acted upon). The latter may include obsolete UIDs that have been superceded by newer UIDs. The obsolete UIDs, as well as any invalid UIDs that were present in your file, are explicitly reported at the top of the Batch Entrez report. You can revise your UID list file on your computer based on that report, if needed, before importing into FLink.
|
|
|


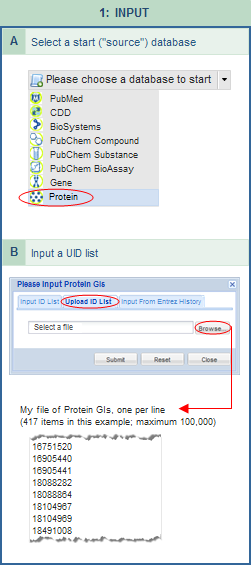
Input summary display: REVIEW input + SELECT destination database  
After you input a UID list directly into FLink, you will see an intermediate display such as the one illustrated here that summarizes your input and allows you to LinkTo a destination database. (If you access FLink by starting with an Entrez Search instead of inputting a UID list directly, you will not see this intermediate display. Any  FLink icons that appear on the Entrez search results page will take you directly to FLink's output display.) FLink icons that appear on the Entrez search results page will take you directly to FLink's output display.)
UID list processed -- After you input a UID list into FLink, the tool checks your list to ensure all the UIDs are valid.
- If all the UIDs are valid, FLink will ignore duplicate UIDs and will open an input summary page such as the one shown in the illustration to the right. Note that it lists the items in order of increasing UID, regardless of the order in which they were input into the system.
- If the input list contains any invalid UIDs, FLink will display an error message (e.g., "Failed to Get List. ERROR:The input file contains invalid UID(s), which cannot be processed by FLink..."). In such a case, remove the invalid UIDs from the input list and submit the job again. (See the tip on using Batch Entrez to check the UIDs in your list.)
Descriptive information displayed -- FLink retrieves descriptive information from Entrez about each valid UID in your list and presents it on the summary page for your review. Note: If your input list contains any UIDs that fall within the acceptable range of integers, 0 to 4294967295, but do not correspond to available database records, these UIDs will appear in the input summary folder tab but no descriptive information will be displayed for them.
Folder tab functions 
As noted in the section on choosing a start database, it is possible to have multiple folder tabs open in the input summary display, one for each starting database. Each folder tab will have the following options:
 Clear Selections -- Clear of the all checkboxes that have been selected on all pages within the folder tab. (Note: It is possible to select items on multiple pages (1 of 10, 4 of 10, 9 of 10, etc.) within a folder tab, and the "clear selections" function will clear the checkboxes on all the pages.) Clear Selections -- Clear of the all checkboxes that have been selected on all pages within the folder tab. (Note: It is possible to select items on multiple pages (1 of 10, 4 of 10, 9 of 10, etc.) within a folder tab, and the "clear selections" function will clear the checkboxes on all the pages.)
 Show records in source database -- Retrieve all (default) or selected items from the corresponding Entrez database. This function will temporarily take you out of FLink and into that Entrez database, but you can use the browser's back button to return to the FLink display. Show records in source database -- Retrieve all (default) or selected items from the corresponding Entrez database. This function will temporarily take you out of FLink and into that Entrez database, but you can use the browser's back button to return to the FLink display.
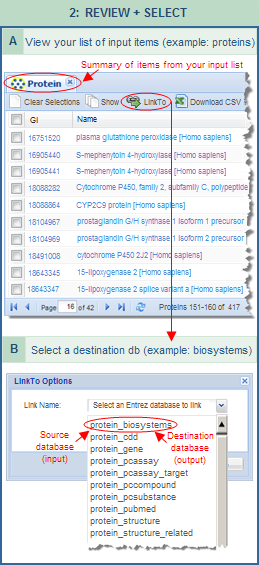
 LinkTo -- This is the main function of the FLink tool, and it acts upon all the records in the folder tab, whether or not their checkboxes are activated. It allows you to select your desired destination database and data type, represented as a "link name." For example, if you are starting in the Protein database, the "Link Name" menu will include choices such as "protein_biosystems," "protein_cdd," "protein_gene," "protein_structure," etc., where the second part of each term indicates the destination database. Some destination databases might have several options, for example, "protein_structure" and "protein_structure_related." To learn the difference, just select an option to read its description in the "LinkTo Options" dialog box. You can continue browsing the various LinkNames in that way, then press the "submit" button in the dialog box once you've selected the LinkName of interest to you. LinkTo -- This is the main function of the FLink tool, and it acts upon all the records in the folder tab, whether or not their checkboxes are activated. It allows you to select your desired destination database and data type, represented as a "link name." For example, if you are starting in the Protein database, the "Link Name" menu will include choices such as "protein_biosystems," "protein_cdd," "protein_gene," "protein_structure," etc., where the second part of each term indicates the destination database. Some destination databases might have several options, for example, "protein_structure" and "protein_structure_related." To learn the difference, just select an option to read its description in the "LinkTo Options" dialog box. You can continue browsing the various LinkNames in that way, then press the "submit" button in the dialog box once you've selected the LinkName of interest to you.
Job ID: After you select a "LinkTo" option and press the "Submit" button, a randomly generated job ID will be displayed in the URL (e.g., https://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi?cmd=getjob&&jobid=237915522). That job ID, and hence the URL, will remain valid for 24 hours and can therefore be used to retrieve the results of a job during that time period.
A separate job ID is assigned to each LinkTo operation. For example, if you import the sample list of 417 protein UIDs into FLink and select "LinkTo: protein_biosystems," the URL that appears on the output page of ranked biosystems will include a randomly generated job ID. Save the URL if you want to retrieve those results within the next 24 hours. If you then want to select another LinkTo option for the same input list, use the browser's back button to return to input summary page and select a new destination option such as "LinkTo: protein_gene." A new job ID for that "LinkTo" operation will appear on the output page, which can also be saved and used over the next 24 hours.
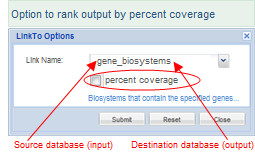 Percent Coverage: Some "LinkTo" options, such as links from the Gene, Protein, and PubChem databases to BioSystems, display a checkbox for "Percent Coverage." If you select the checkbox, the FLink output will rank output by that value rather than by "Frequency." The "Frequency" column will still be present in the output and can still be used to re-sort the data by that value, if desired. Details about both sorting methods are provided in the section on "ranked list of records." Percent Coverage: Some "LinkTo" options, such as links from the Gene, Protein, and PubChem databases to BioSystems, display a checkbox for "Percent Coverage." If you select the checkbox, the FLink output will rank output by that value rather than by "Frequency." The "Frequency" column will still be present in the output and can still be used to re-sort the data by that value, if desired. Details about both sorting methods are provided in the section on "ranked list of records."
(Illustrated examples of pathways ranked by percent coverage vs. pathways ranked by frequency are provided in the "How To" file on "Start with a gene expression study and retrieve a ranked list of pathways (biosystems) in which the up- or down-regulated genes are involved.")
 Download CSV file -- Download the data displayed in the folder tab as a comma separated value (CSV) file. This function acts upon all the records in the display, whether or not their checkboxes are activated. Download CSV file -- Download the data displayed in the folder tab as a comma separated value (CSV) file. This function acts upon all the records in the display, whether or not their checkboxes are activated.
Tip: The csv file is in Unicode (UTF-8) format. If you plan to import the file into a spreadsheet program, you might need to specify the Unicode (UTF-8) format during the import process. For example, if you plan to import the file into Excel, follow the instructions at Tune Help (https://help.tune.com/marketing-console/how-to-import-a-unicode-csv-to-excel/), starting with step 4, in order to import Unicode properly. Note: If the Unicode (UTF-8) format is not specified during the process of importing the CSV file into the spreadsheet program, certain ASCII characters such as the umlaut (ö), e-acute (é), etc. might not appear correctly.
 Summary -- Display additional descriptive information for all records, as available, whether or not their checkboxes are activated. Summary -- Display additional descriptive information for all records, as available, whether or not their checkboxes are activated.
Close folder tab -- The "X" in the upper right hand corner of a folder tab on the input summary display will close that tab and will delete from the server the UID list it currently contains. The database tab can be opened again later, if desired, by selecting it from the start menu. A new UID list can then be imported.
Start Over -- Pressing the "Start Over" button at the bottom of the input summary display will close all of the folder tabs and will delete from the server the UID lists they currently contain. If you prefer to close only one of the "start" databases and delete its UID list, simply close the appropriate folder tab.
View output  
|
 
A second way to use FLink is to start by searching an Entrez database of interest (from the list of supported databases) and then link to FLink from the search results page. The name an appearance of the link might vary. For example:
If you start a search in the GEO Profiles database, the search results will display a "Find Pathways" button in the right-hand margin of the page. That button performs a multiple-step FLink operation which retrieves a ranked list of pathways associated with the gene expression studies. (A detailed example is provided in the "How To" file on "Start with a gene expression study and retrieve a ranked list of biosystems in which the up- or down-regulated genes are involved."
If you start a search in some other databases, the link to FLink will appear on the search results page as an  icon and will retrieve the related data from the destination database as a ranked list. icon and will retrieve the related data from the destination database as a ranked list.
Important notes about displays that use the  FLink icon: FLink icon:
- clicking on the link name (e.g., "BioSystems") that appears beside the icon will open the records directly in the destination database, where records will be listed in the default sort order for that database. In contrast, clicking on the FLink icon (
 ) will display the records as a frequency-weighted list in the FLink tool. ) will display the records as a frequency-weighted list in the FLink tool.
- Only a subset of supported databases currently contain
 FLink icons on their search results page at the time of FLink's initial release. If you do an Entrez search in a supported database and the search results page does not display any FLink icons, you can still access frequency-weighted lists of related data for your search results. This is because your search results are temporarily stored in Entrez History (for up to 8 hours after your last activity in a given Entrez database) and they can be imported as a UID list directly into FLink. To do this, open the FLink icons on their search results page at the time of FLink's initial release. If you do an Entrez search in a supported database and the search results page does not display any FLink icons, you can still access frequency-weighted lists of related data for your search results. This is because your search results are temporarily stored in Entrez History (for up to 8 hours after your last activity in a given Entrez database) and they can be imported as a UID list directly into FLink. To do this, open the  FLink home page, choose the database you just searched as the starting point, then select "Input from Entrez History" in the dialog box. Your search results will be imported into FLink and you can then use the "LinkTo" menu to select the desired destination database and data type, as described above. FLink home page, choose the database you just searched as the starting point, then select "Input from Entrez History" in the dialog box. Your search results will be imported into FLink and you can then use the "LinkTo" menu to select the desired destination database and data type, as described above.
 
 FLink can accept up to 100,000 UIDs as input (and can display up to 100,000 associated UIDs from the destination database on the output page). FLink can accept up to 100,000 UIDs as input (and can display up to 100,000 associated UIDs from the destination database on the output page).
|