- Use the CD-Search tool to compare a protein query sequence (either as raw sequence data in FASTA format, or as a GI or Accession) against the desired conserved domain data set, in order to identify functional units within the query sequence. The search results will be shown in the default Concise Display, which shows only the top scoring hits for each region of the query sequence.
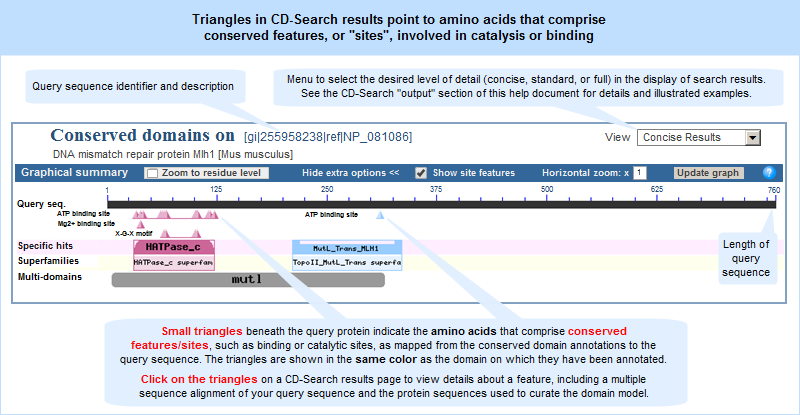
- The small triangles beneath the query protein on a CD-Search results page indicate the residues that comprise conserved features/sites, such as binding or catalytic sites, as mapped from the conserved domain annotations to the query sequence. The triangles are shown in the same color as the domain on which they have been annotated and appear if a region of the query protein sequence: (1) gets a specific hit to a domain model on which conserved features/sites have been annotated, OR (2) gets a non-specific hit to a domain model that belongs to a superfamily whose representative is an NCBI-curated domain that has such annotations. (These hit types are shown on illustrations of the CD-Search results Concise Display and Full Display.)
- Click on the triangles to view details about the feature, including a multiple sequence alignment of your query sequence and the protein sequences used to curate the domain model. Hash marks (#) above the aligned sequences show the location of the conserved feature residues (see illustration in help document). A thumbnail image, if present, provides an approximate view of the feature's location in three dimensions and options for interactive 3D structure viewing.
| |
The example above shows the search results, as of October 22, 2014, for protein GI 255958238 (mouse DNA mismatch repair protein Mlh1). Click anywhere on the graphic to view the actual, interactive CD-Search results page.
The hit types in the full display can include specific hits, non-specific hits, the superfamily(ies) to which those hits belong, and multi-domain models. Each superfamily is represented by a cartoon with a distinct color/shape combination, in order to distinguish domains from each other. If conserved features/sites are present, triangles are shown all three types of search results displays (concise results, standard results, and full results).
|
|
|