| Related Structures (CBLAST) Help Document |
|
|
| |
This help document describes the Related Structures resource (also known as CBLAST), which accepts a protein sequence as input, and then finds experimentally resolved 3D structures that are related to the query protein based on sequence similarity. (Note: A separate resource, the Vector Alignment Search Tool (VAST), identifies similar protein 3-dimensional structures by purely geometric criteria, and can be used if your query is a protein structure rather than a protein sequence.) |
|
| |
|
|
| |
| |
The Related Structures service accepts a protein sequence as input, and then finds experimentally resolved 3D structures that are related to the query protein based on sequence similarity. These are referred to as related structures, and can be used to infer the structure of the query protein, and identify/predict functional sites based on the alignment information, even if the query protein does not yet have an experimentally resolved structure itself.
To do this, the Related Structures service uses BLAST to compare the protein query sequence against the protein sequences from all structures in the Molecular Modeling Database (MMDB) in pairwise comparisons. It then lists statistically significant matches, sorted by similarity scores.
This approach is used because the 3D structure (tertiary structure) of a protein is considered to be largely determined by the its amino acid sequence (primary structure). Therefore, if the amino acid sequence of a protein is similar to that of a protein whose 3D structure is known, we can then assume that the query sequence is likely to have a similar 3D structure.
The Related Structures service is also referred to as "CBLAST," where "BLAST" represents the sequence similarity search tool that is used to find proteins (from experimenally resolved structures) that are related to the query protein, and the "C" represents the Cn3D structure viewer that can be used to interactively view the 3D protein structures that are found along with the sequence alignment suggested by BLAST, and to map aligned regions to the 3D structure space.
(NOTE: A separate service, the Vector Alignment Search Tool (VAST), identifies protein 3-dimensional structures that are similar to each other by purely geometric criteria. These are referred to as similar structures, and the VAST help document provides additional details about that tool.)
|
|
|
|
|
| |
| |
A 3D structure can often provide detailed information on a protein's biological function and mechanism of action, but experimentally solving a 3D structure is no easy task and is not always possible. Therefore, only a small fraction of known proteins have their 3D structure information available.
For other proteins, however, some structural information may be inferred by comparison to 3D structures of proteins that are in the same sequence family (based on sequence similarity) -- that is, by examination of related structures.
The Related Structures service finds 3D protein structures that are similar in sequence to your query protein. It presents a 3D view of each related structure together with a pairwise alignment of the query protein sequence and the 3D structure's protein sequence.
The Related Structures service also searches our Conserved Domain Database (CDD) to identify conserved domains in the query protein sequence, and to map functional sites from the conserved domains onto the query sequence.
Each related structure and pairwise sequence alignment can be downloaded and viewed in Cn3D, or saved on a local computer for later use.
The following pages provide illustrated examples that demonstrate the use of Related Structures, using the human prostaglandin-endoperoxide synthase 1 isoform 1 precursor (accession NP_000953.2, gi 18104967), which is a product of the human PTGS1 gene (GeneID 5742), as the protein query sequence:
|
|
|
|
|
| |
|
|
| |
Direct search in the Related Structures service, using a protein GI number as the query:
 
The Related Structures home page enables you to enter the sequence identification number (i.e., the GI number) of any protein sequence that is available in the Entrez Protein database, and retrieves related structures (as available), which have been pre-computed for quick retrieval. For example:
- Open the Related Structures home page.
- In the text box, enter 463989, which is the GI number for protein accession AAC50285: DNA mismatch repair protein homolog [Homo sapiens].)
- Press the "Find related structures" button to retrieve proteins that are similar in sequence to your query, and that have experimentally resolved structures. (View the related structures for this sample query.)
Link from an Entrez Protein sequence record to Related Structures:
 
Entrez Protein sequence record displays also provide access to Related Structures, by displaying a "Related Structures" link in the right margin of protein sequence record displays. For example:
- Open the protein sequence record AAC50285 (GI 463989), for the human DNA mismatch repair protein homolog, in the Entrez Protein sequence database and scroll down the page. In the right-hand margin, you will see a "Related Information" section, which includes links for "Related Structures (list)" and "Related Structures (summary)." The latter link opens a page with a graphical display that summarizes conserved domains and conserved features/sites found on the protein query sequence, the alignment footprints of related structures, and links that allow you to display the 3D structure and sequence alignment in Cn3D.
(These steps are illustrated below, where the protein sequence record is shown in FASTA format. The "Related Information" links also appear in the right hand margin when the sequence record is displayed in other formats, including the default GenPept format.)
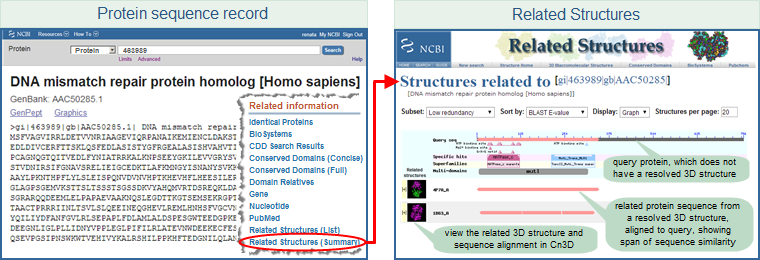
- The Molecular Modeling Database (MMDB) help document provides more information about the "Structure", "Related Structures (List)", and "Related Structures (Summary)" links that are accessible (as available) from the right margin of protein sequence record displays.
- The following pages provide illustrated examples that demonstrate the use of the "Related Structures" link, using the human prostaglandin-endoperoxide synthase 1 isoform 1 precursor (accession NP_000953.2, gi 18104967), which is a product of the human PTGS1 gene (GeneID 5742), as the protein query sequence:
Protein BLAST against the PDB data set, using a protein GI number as the query:
 
Protein BLAST search results also provide access to Related Structures, by displaying a "Structure" link in the right margin of a pairwise sequence alignment on a protein BLAST results page, if a BLAST hit is from the Protein Data Bank (PDB). For example:
- Open the Protein BLAST query page, and enter 463989 as the protein query sequence (463989 is the GI number for the human MLH1 protein homolog). In the "Choose Search Set" section of the query page, select "Protein Data Bank proteins (pdb)," and press the "BLAST" button near the bottom of the page to start the search. On the BLAST results page for GI 463989, click on the description of any hit to view a pairwise alignment between the protein query sequence and the BLAST hit. Each pairwise alignment will show a "Related Information: Structure" link in the right margin of the display, because all of the BLAST hits are from the Protein Data Bank, which we chose as the search set, and therefore have a 3D structure.
- Note: If you choose the default "nr" (non-redundant) database (instead of the "Protein Data Bank proteins (pdb)") in the "Choose Search Set" menu, then only the hits that have 3D structures will show the "Related Information: Structure" link in the right margin of their pairwise alignment. If you do not see a "Related Information: Structure" link in the right margin of a pairwise alignment, that means the BLAST hit is not from a 3D structure record.
Protein BLAST against the PDB data set, using protein sequence data (in FASTA format) as the query:
 
If your query protein is not yet publicly available in the Entrez Protein database (and therefore does not yet have a GI number), you can still find related structures by doing a BLAST search of your query protein sequence data against the protein sequences from the Protein Data Bank (PDB), each of which has an experimentally resolved 3D structure. To do this:
- Open the protein BLAST search page
- In the "Enter Query Sequence" section of the page, type/paste your query protein sequence data (preferrably in FASTA format) into the text field box.
- In the "Choose Search Set" section of the page, select "Protein Data Bank proteins(pdb)" as the database to search against.
- Click on the "BLAST" button near the bottom of the page to start the search.
- After the BLAST search is completed, click on a hit of interest to view its pairwise sequence alignment with the query sequence. Look for the "Structure" link in the right margin of the pairwise sequence alignment display. Click on the "Structure" link and the Related Structures service will open in a new window/tab.
|
|
|
|
|
| |
| |
Below is an example of a Related Structures search results page, for the query protein sequence GI 257051069, transitional endoplasmic reticulum ATPase from Xenopus laevis. The query sequence, which does not have an experimentally resolved structure, is aligned to similar protein sequences that do have experimenally resolved 3D structures. (Click on the image to open the live Related Structures search results page for GI 257051069.)
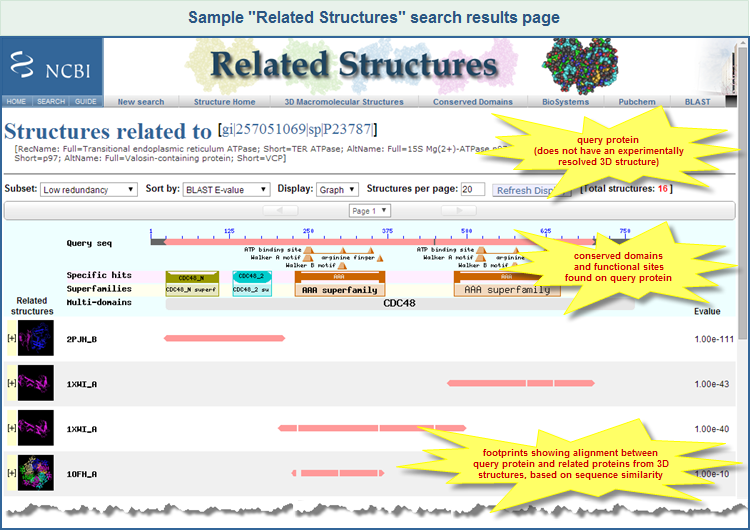
The top of the display summarizes information about your query sequence, including its GI number, Accession, and definition line (description). The sequence identifiers are shown in FASTA defline format (e.g., [gi|nnnnnn|db|XXXXXX], where nnnnnn is the GI number, db is an abbreviation for the source database (such as "sp" for Swiss Prot), and XXXXXX is the accession number). The sequence identifiers link to the corresponding sequence record in the Entrez Protein protein database, enabling you to open the sequence record, if desired. (Note: If you entered a protein query sequence that is not yet available in the Entrez Protein database, then the top of the display will show the sequence identification information that you provided in your FASTA-formatted query sequence.)
In the graphic display, the ruler labed "Query Seq" represents the query sequence with amino acid positions labeled, providing a defined scale for the alignments. Below the ruler are small triangles that indicate conserved features/sites, and cartoons with distinct colors/shapes that indicate conserved domains. These were found in the query sequence by the CD-Search service, which uses RPS-BLAST to compare a query protein sequence against the Conserved Domains Database (CDD). A conserved domain that appears on the line labeled "Specific hits" indicates a high confidence that the query sequence belongs to the same protein family as the sequences used to create the domain model. (Please refer to the Conserved Domain Database Help document, and the CD-Search Help document, for more information about those resources, including types of hits such as specific, non-specific, superfamily, and multidomain).
Related structures are displayed below conserved domains panel. Each structure occupies an entire row that contains:
- A thumbnail of the structure, with an option to interactively view the structure and sequence alignment in Cn3D
- A PDB-style sequence ID of the related structure
- An alignment footprint (pink line) that shows the region of sequence similarity between the query protein and the related structure
- The BLAST score (E-value (default), bit score, alignment length, sequence identity) that is used to sort the related structures.
To view more details about a related structure, click on the row that lists the structure of interest. That will toggle open/close a panel that provides additional information, such as:
- A larger model picture of the structure
- A link to the corresponding structure record in the Molecular Modeling Database (MMDB), where all structures are stored, and where more details about each structure are available
- A link to search references (publications) of the structure in PubMed database
- The description title of the structure
- All four BLAST scores (E-value, bit score, alignment length, and sequence identity) for the alignment between the query protein sequence and the 3D structure's protein sequence. (Details about those scores, and other terms related to sequence similarity searching, are provided in the BLAST Glossary and NCBI Handbook Glossary.)
- A pairwise sequence alignment, illustrated below, provides a detailed, residue-by-residue comparison of the query protein and the 3D structure's protein:
- identical residues are in shown red
- similar residues in blue
- non-matched residues in grey
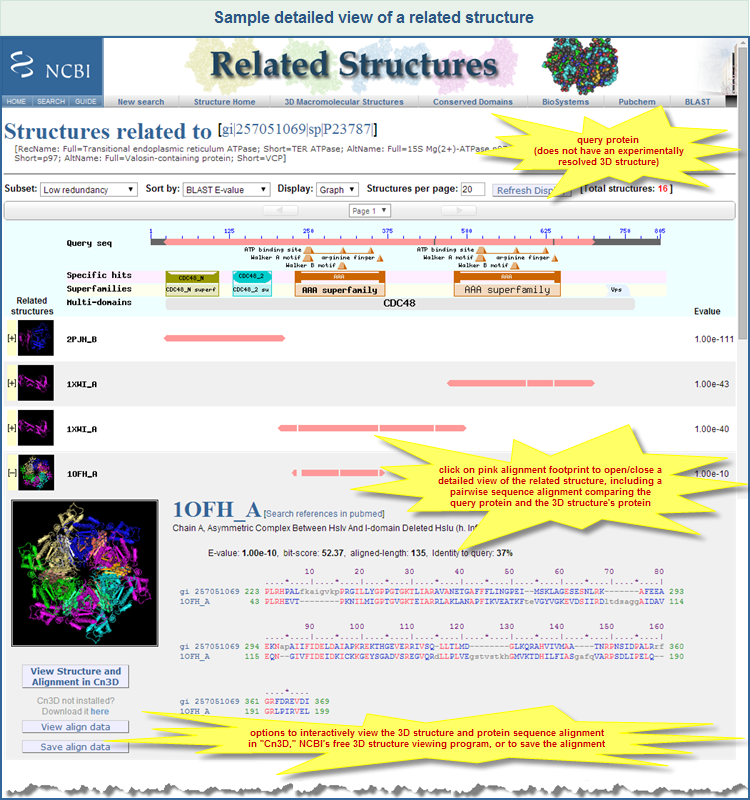
The detailed view panel also provides three buttons (below the model picture) that enable you to view or download the structure/alignment data:
- Clicking on
 to download the data and display them in Cn3D. (Cn3D must be installed on your computer in order for the button to work. A tutorial shows how the progam can be used.) to download the data and display them in Cn3D. (Cn3D must be installed on your computer in order for the button to work. A tutorial shows how the progam can be used.)
- Clicking on
 will download the data in a human-readable format (ASN-text) and present them in the browser window. will download the data in a human-readable format (ASN-text) and present them in the browser window.
- Clicking on
 will download the data in binary (ASN-binary, not human-readable) and prompt you to save the file on local computer. will download the data in binary (ASN-binary, not human-readable) and prompt you to save the file on local computer.
Both ASN-text and ASN-binary formats are recognized by Cn3D, regardless of file name or extension. If you save the data in a file with a .cn3 extension, in the Windows operating system, you should be able to open the file with Cn3D by double-clicking.
If there are too many structures found, the results may be paginated. There are two navigation bars (one bar above the graphic display and an identical bar beneath the graphic display, just for convenience) that allow you to page through the results using the left and right arrows, or to jump to a specific page by selecting it from the drop-down menu that appears between the arrows.
|
|
|
|
|
| |
| |
There is a control panel, above the graphic display of the query sequence, that contains several options that enable you to choose how the result should be displayed:
- The "Subset" menu allows you to select the level of redundancy that you would like to see in the display of search results. (The default setting is "Low redundancy.") A separate section of this document provides additional information about redundancy levels, and the method used for clustering structures in order to provide various levels of redundancy in search results.
- The "Sort by" menu allows you to select which similarity score (E-value, bit score, alignment length, sequence identity) should be used to sort the results.
- The "Display" menu enables you to view the results either as a:
- Graphic summary ("Graph," the default setting), which shows the alignment footprints (pink bars) of the related structures relative to the query protein (illustrated example). It also provides a detailed view that shows the pairwise sequence alignment of the query protein and the related structure's protein (illustrated example), along with options to view the 3D structure and sequence alignment in Cn3D. (The detailed view is accessible by clicking on the "+" beside the thumbnail graphic of the related structure, or by clicking on the pink alignment footprint.)
- "Table," which shows the thumbnail molecular graphic, structure identifiers (PDB ID and MMDB ID), description, and BLAST scores (E-value, bit score, alignment length, sequence identity) for each related structure. (The Table display also enables you to save the results for future reference; simply select/copy/paste the desired subset of results into your preferred file type (e.g., *.txt, *.doc, spreadsheet.)
- The "Structures per page" text box enables you to specify how many structures should be listed on one page.
After everything is selected, click on the "Refresh Display" button.
|
|
|
|
|
| |
| |
The "Subset" menu on a Related Structures search results page allows you to select the level of redundancy you want to view in the display.
Many proteins may have identical or very similar amino-acid sequences (for example, several proteins sequenced from different organisms by different labs may turn out to have identical amino-acid sequences). Search results that display every one of those sequences can be redundant.
To address this, the structures in MMDB are clustered into groups based on protein sequence similarity. Structures in each group are ranked according to apparent quality and completeness of the structure data, and only the highest ranked structure will be listed as a representative of the group, thus reducing redundancy in the display of related structures.
The available levels of redundancy are listed below, and are determinued by the E-value threshold used for clustering. A smaller E-value threshold means more strict clustering, i.e., fewer structures are considered to be similar and clustered into one group, which results in more groups and therefore more redundancy in the search result. You can choose from five redundancy levels:
- All similar MMDB -- No clustering. All related structures are listed. This is the highest redundancy level.
- Non-identical -- Only identical sequences are grouped into a cluster, and one representative from each cluster is shown in the results. Very high redundancy.
- High redundancy -- Proteins are clustered based on sequence similarity using an E-value threshold of 10-80, and one representative from each cluster is shown in the results.
- Medium redundancy -- Proteins are clustered based on sequence similarity using an E-value threshold of 10-40, and one representative from each cluster is shown in the results.
- Low redundancy -- Proteins are clustered based on sequence similarity using an E-value threshold of 10-7 (default), and one representative from each cluster is shown in the results.
The Vector Alignment Search Tool (VAST), which identifies similar protein 3-dimensional structures by purely geometric criteria (not by sequence similarity), also uses this type of clustering to present search results. The VAST help document provides additional details about the method used for clustering.
Note that the redundancy levels are provided as a convenience, in order to make browsing Related Structures search results faster and easier. Nevertheless, even when a cluster contains identical protein sequences, there might be interesting variations among members of the cluster. For example, some might be free proteins while others might be bound to another molecule. If such variations are of interest, select "All similar MMDB" from the "Subset" menu to view the complete set of search results.
|
|
|
|
|
| |
| |
If the Related Structures service does not find any hits for a your query protein sequence (or if there are no "Related Structures" links available in the right hand margin of an Entrez Protein sequence record display), the following approaches can help you to find more distantly related structures:
- PSI-BLAST
 
Position-Specific Iterated BLAST (PSI-BLAST) can find more distantly related proteins than the regular protein BLAST program, and some of the more distantly related proteins might be associated with structures. The first iteration of PSI-BLAST search results might not contain any protein sequences derived from 3D structure records, but subsequent iterations will find more distantly related proteins, some of which might have experimentally resolved 3D structures. If a PSI-BLAST hit is associated with a 3D structure, it will have a "Structure" link in the right hand margin of the pairwise alignment of the query sequence and the PSI-BLAST hit.
To use PSI-BLAST:
- Open the protein BLAST (blastp) page.
- Select the search parameters:
In the "Search Set" section of the page, select "non-redundant protein sequences (nr)."
In the "Program Selection" section, and select "PSI-BLAST (Position-Specific Iterated BLAST)." (Or simply click on the link at the beginning of this paragraph, which will open the protein BLAST page with those search parameters already selected.)
- Enter the protein query sequence as a GI number or as FASTA-formatted sequence data.
- Press the "BLAST" button at the bottom of the page.
Read more about PSI-BLAST in:
- CD-Search
 
The CD-Search service is a web-based tool for the detection of conserved domains in protein sequences. It can therefore help to elucidate the protein's function. Many conserved domains, particularly NCBI-curated domain models, are based on multiple sequence alignments that include proteins from experimentally resolved 3D structures. Therefore, if the CD-Search service finds conserved domains in your query sequence, and if some of the hits are NCBI-curated domain models (or members of conserved domain superfamilies associated with 3D structures), it is likely that you can see 3D structures that are related to the functional parts of your query sequence, even if the Related Structures (CBLAST) service did not find hits for your overall query protein.
To use CD-Search:
- Open the CD-Search page.
- Enter the protein query sequence as a GI number or as FASTA-formatted sequence data.
- Adjust the options (search parameters), if desired.
- Press the "Submit" button at the bottom of the page.
- On the search results page (illustrated example) , you can spot NCBI-curated domain models because they have a "cd" accession number prefix (e.g,. cd00400. They also may appear as specific hits. If the conserved domain is from another source database, the superfamily to which it belongs might be associated with a 3D structure.
- Click on the colored graphic for a domain model (or superfamily) of interest to view detailed information in the Conserved Domain Database. The detailed view includes a multiple sequence alignment of the proteins used to curate the domain model, with your query protein embedded in the alignment. It also may include, if/as available, thumbnail images and/or links to 3D structures of the domain.
- Clicking on the thumbnails will open the 3D structure in the free Cn3D structure viewing program (if that program is already installed on your computer), along with an alignment of the proteins used to curate the domain.
- Clicking on the "Structure View" button in the left hand margin will open a similar view, but your query protein will also be present in the multiple sequence alignment.
Read more about CD-Search in the articles that are accessible from the:
|
|
|
|
|
| |
| |
 |
Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, Zhang N, Bryant SH. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007 Jan; 35(Database issue): D298-300. [PubMed PMID: 17135201] [Full Text] |
|
| |
|
(See all publications about NCBI's 3D Macromolecular Structures Resources, including the one listed here plus articles by the NCBI Structure group describing the results of computational biology research on the Molecular Modeling Database.) |
|
|
|
|
|
|



 to download the data and display them in
to download the data and display them in  will download the data in a human-readable format (ASN-text) and present them in the browser window.
will download the data in a human-readable format (ASN-text) and present them in the browser window.
 will download the data in binary (ASN-binary, not human-readable) and prompt you to save the file on local computer.
will download the data in binary (ASN-binary, not human-readable) and prompt you to save the file on local computer.